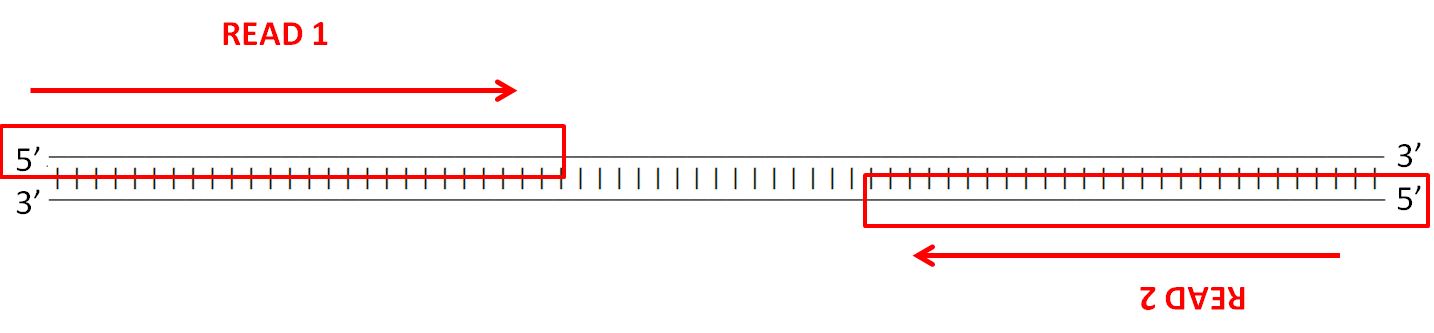
paired end sequencing reads
Paired-end sequencing allows users to sequence both ends of a fragment and generate high-quality alignable sequence data. Visit Maverix Biomics to learn more about RNA-seq.
How Sequencing Works Ngs Analysis
When you align them to the genome one read should align to the forward strand and the other should align to the reverse strand at a higher base pair position than the first one.
. The Illumina paired-end sequencing technology can generate reads from both ends of target DNA fragments which can subsequently be merged to increase the. For sequencing projects that require higher accuracy such as studies of alternate splicing 40 million to 60 million paired-end reads will provide better results. Biocc paired end or mate pair refers to how the library is made and then how it is sequenced.
Paired-end sequencing facilitates detection of genomic. This can be done using the Set Paired Reads. In general paired-end reads tend to be in a FR orientation have relatively small inserts 300 - 500 bp and are particularly useful for the sequencing of fragments that.
Paired-end DNA sequencing reads provide high-quality alignment across DNA regions containing repetitive sequences and produce long contigs for de novo. Both are methodologies that in addition to the sequence information give you. The standard Illumina paired-end protocol produces reads oriented pointing toward each other just like good old fashioned Sanger paired reads but the insert size is much.
Paired-end 150 means that one read of 150 bases in size is generated from each end of the fragment through the inserted middle piece of target DNA from both directions for a total of 2. With paired-end sequencing after a DNA fragment is read from one end the process starts again in the other direction. Mate pair sequencing is used for various applications applications including.
Illumina sequencing of a marker gene is popular in metagenomic studies. Another supposed advantage is that it leads to more accurate. Read files from paired-end sequencing need to be paired in Geneious before the pairing information can be used in assembly.
In addition to producing twice the number of sequencing reads this. Paired-end sequencing facilitates detection of genomic rearrangements and repetitive sequence elements as well as gene fusions and novel transcripts. RNA-seq analysis configuration on the Maverix Analytic Platform.
Paired-end DNA sequencing reads provide high-quality alignment across DNA regions containing repetitive sequences and produce long contigs for de novo sequencing by filling gaps in the. Combining data from mate pair sequencing with those from short-insert paired-end reads provides increased. One of the advantages of paired end sequencing over single end is that it doubles the amount of data.
For more detailed analyses to. Since paired-end reads are more likely. Paired-end vs single-end sequencing reads.
Paired-end DNA sequencing reads provide high-quality alignment across DNA regions containing repetitive sequences and produce long contigs for de novo sequencing by filling gaps in the. However Illumina paired-end PE reads sometimes cannot be merged into single reads for.

Paired End Mapping Reveals Extensive Structural Variation In The Human Genome Science
How Sequencing Works Ngs Analysis

Long Fragments Achieve Lower Base Quality In Illumina Paired End Sequencing Biorxiv
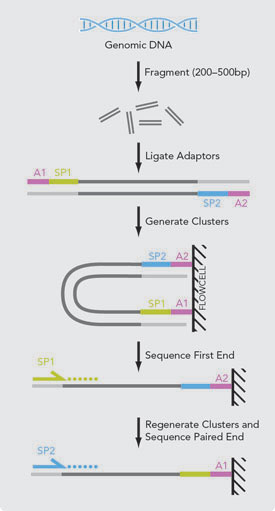
Paired End Sequencing Vs Mate Pair Sequencing Zhongxu Blog
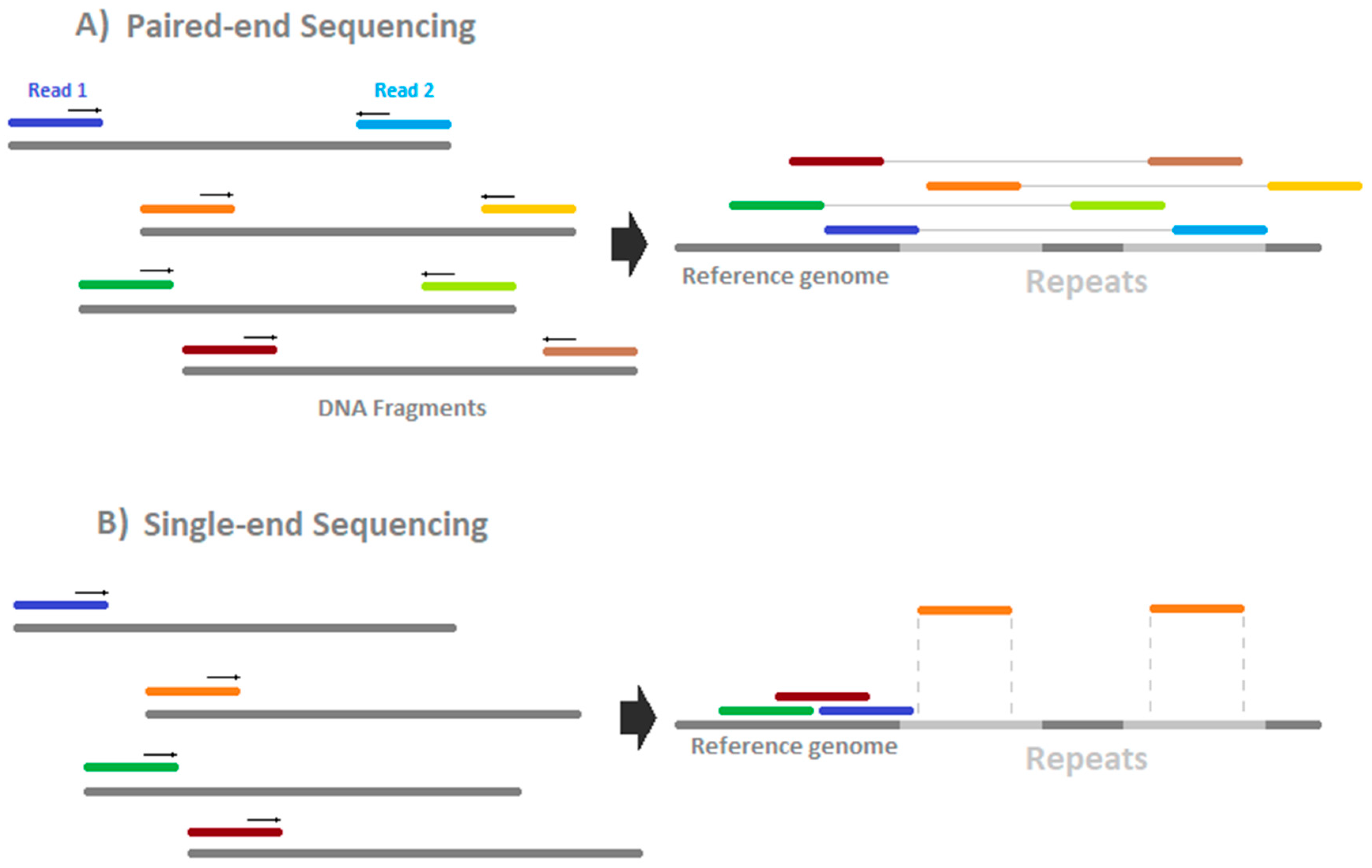
Epigenomes Free Full Text How To Design A Whole Genome Bisulfite Sequencing Experiment Html
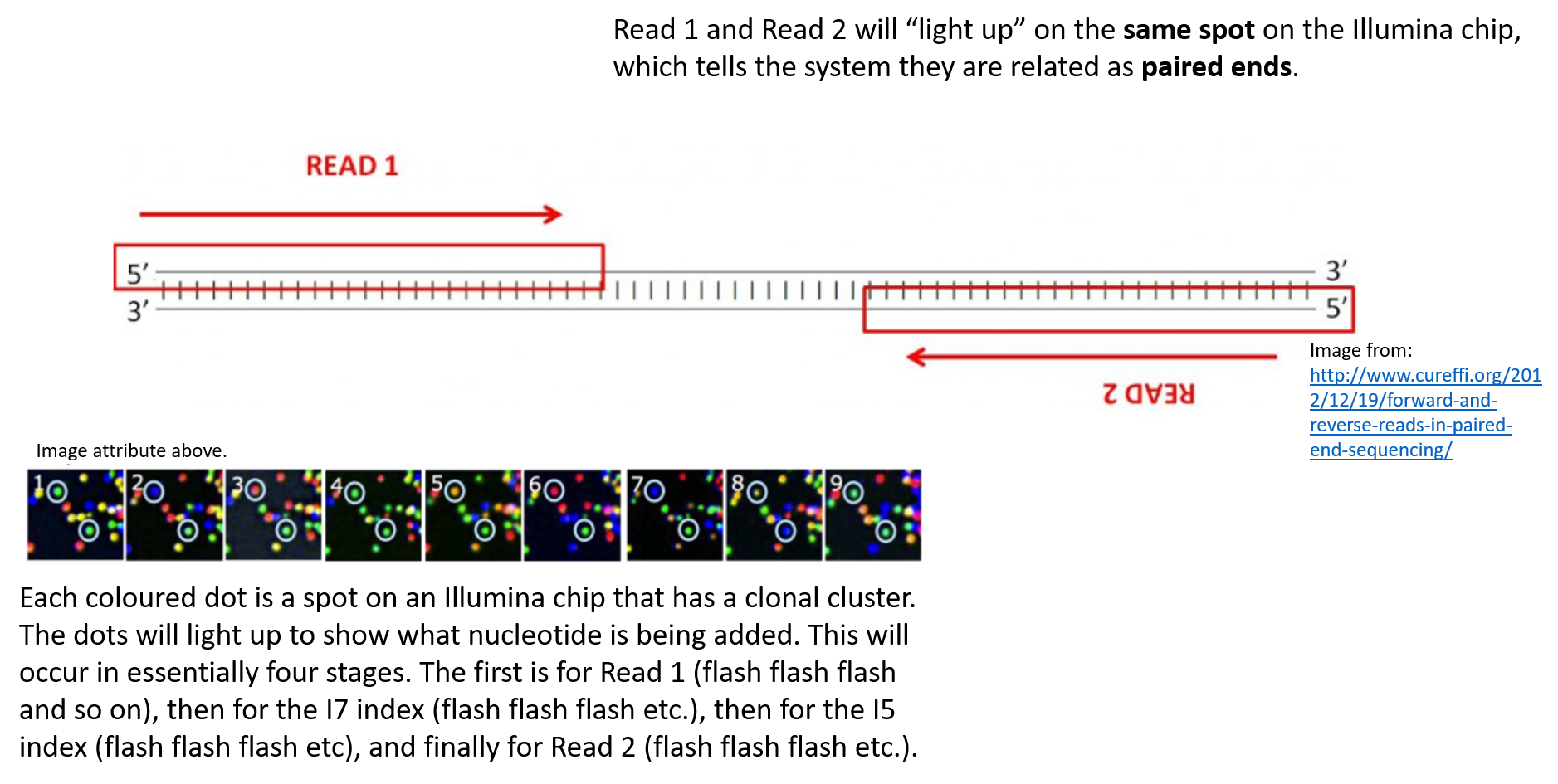
Illumina Sequencing For Dummies An Overview On How Our Samples Are Sequenced Kscbioinformatics
How Short Inserts Affect Sequencing Performance
Paired End V S Single End 有勁的基因資訊 痞客邦
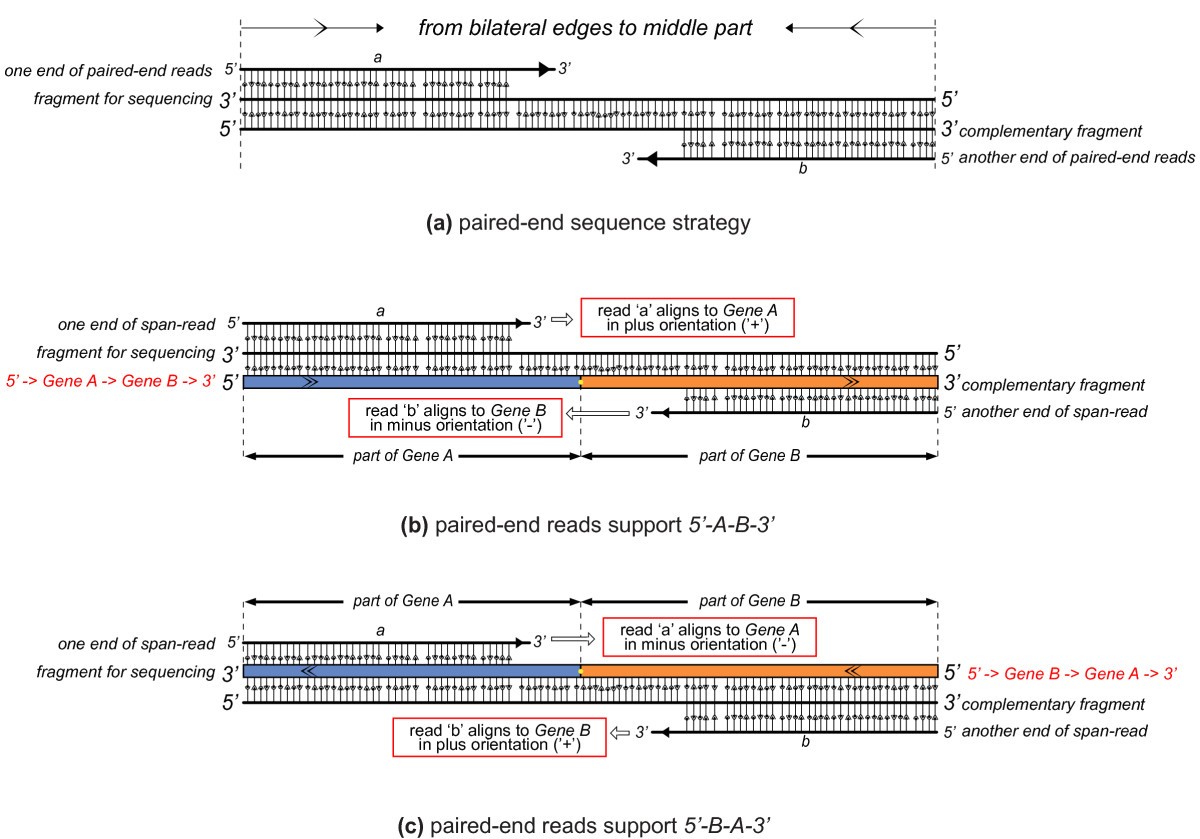
Soapfuse An Algorithm For Identifying Fusion Transcripts From Paired End Rna Seq Data Genome Biology Full Text
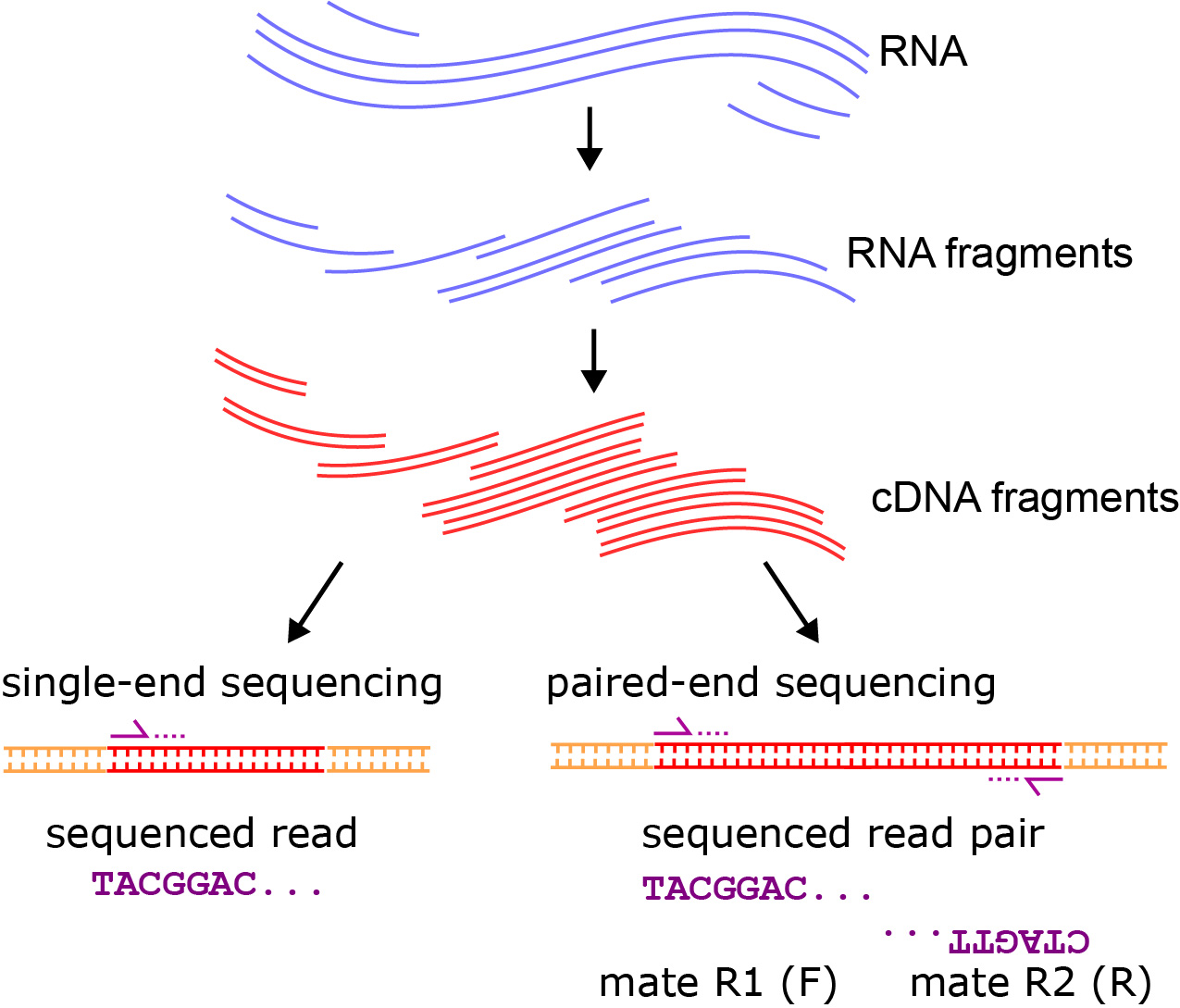
Chapter 6 Transcriptomics Applied Bioinformatics

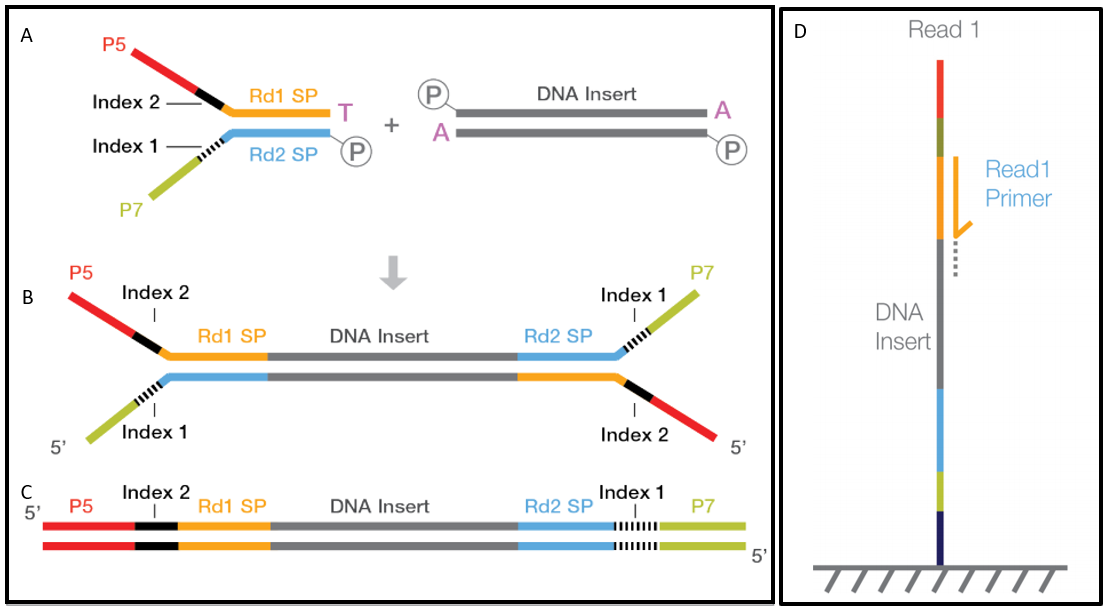
Paired End Sequencing Left Showing Read 1 And Read 2 Primers Starting Download Scientific Diagram

Illumina Sequencing Illumina Sequencing By Synthesis 1010genome Quality Ngs Bioinformatics Data Analysis Services

Paired Vs Unpaired Dna Sequencing

Mapping And Further Processing Gbprocess 4 0 0 Documentation

Chimeric Transcript Discovery By Paired End Transcriptome Sequencing Pnas

Rna Seq How To Know Which Paired Reads Come From The Same Original Fragment
Illumina Index Sequencing Where Is My Sample Enseqlopedia

Beyond The Linear Genome Paired End Sequencing As A Biophysical Tool Trends In Cell Biology